Reproducibility is at the core of solid scientific and technical research. The ability to repeat the research that is produced is a key approach for confirming the validity of a new scientific discovery. Ensuring the robustness and trustworthiness of science that is done using computing and data is critical. The inability to reproduce the results of an experiment can lead to a range of consequences, including the retraction of research and injury to the reputations of the authors and the conference / journal that published it. Furthermore, fostering reproducibility facilitates dissemination, comparison, and adoption of research. Therefore, it can also contribute to increasing the impact and enhancing the visibility of articles.
The International Conference on Parallel Processing (ICPP) recognizes the importance of fostering and enabling reproducible research and it is also committed to raising standards and good practices for reproducibility. For that reason, ICPP is converging with other similar reproducibility initiatives such as the IEEE Transactions on Parallel and Distributed Computing Journal (TPDS) and The International Conference for High Performance (SC’23) reproducibility initiatives, while maintaining the inherent characteristics of the conference.
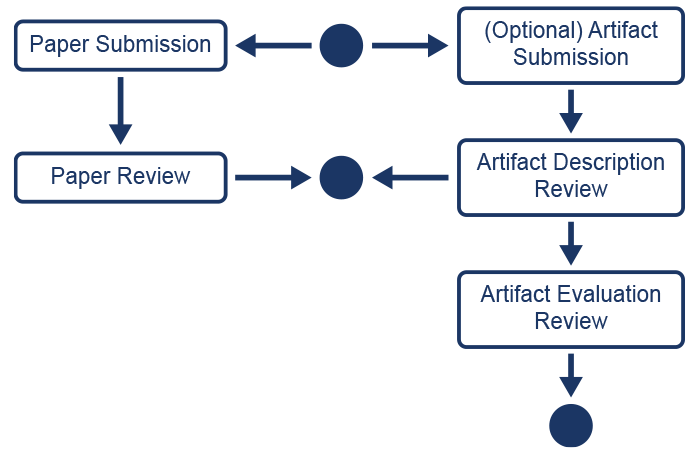
In parallel to the submission of a paper, ICPP authors can optionally submit their reproducibility artifacts that enable reviewers to reproduce the experiments of their papers (Fig. 1). Unlike ICPP paper review, which is double-blinded, ICPP is piloting a single-anonymous peer review of artifacts. The reproducibility committee will not share any kind of information with the paper committee that can compromise the double-blinded review process. As a result of the reviewing process, reproducibility badges (the badges granted as a result of the evaluation of the artifact(s)) may be granted.
As depicted in Figure 1, the reproducibility review process consists of two stages:
- The Artifact Description review stage (in parallel with paper review)
- The Artifact Evaluation review stage (after paper acceptance)
During the artifact description review stage, reviewers will check that the experiments in the artifact are properly documented, and that they include a complete description of the expected results and an evaluation of them, and most importantly how the expected results from the experiment workflow relate to the results found in the paper.
Once the review stage for papers is complete and the accepted papers are determined, the artifact evaluation stage will commence. Only the accepted papers that submitted their reproducibility artifacts will be considered. As a result of this stage and the full evaluation of reproducibility, badges (the badges granted as a result of artifact(s) evaluations) may be granted.
There are four key aspects in reproducibility in practice:
- The computational artifact(s).
- The description (documentation metadata) of the artifact(s).
- The computational resources required to enable reproducibility.
- The badges granted as a result of the evaluation of the artifact(s).
Reproducibility Process for Authors
These are the steps that authors need to consider for the ICPP reproducibility initiative. Maximizing automation throughout the entire process is highly desirable. In this context, we kindly ask authors to provide all necessary scripts for deploying and installing their computational artifacts, particularly at the Artifact Evaluation (AE) stage. The overarching objective is to facilitate the reproducibility of the paper's experiments, allowing for the potential reuse of results by other researchers. A common obstacle to experiment reproducibility lies in intricate installation, deployment, and configuration setups, which often demand substantial efforts from third-party researchers and practitioners.
AD phase – Fast-check reproducibility examination
- Data Artifact Documentation. Then, for the ICPP24, the dataset must be documented. It can include methodological aspects about how the dataset was obtained, if there was any preprocessing and transformation, etc. More information is available in the computational artifact(s).
- Code Artifact Hardware Dependencies. Specify and document hardware requirements (including type and quantity). It could be the case that the code artifact does not have any requirements, then it should be stated. In some cases, the requirements can involve specific CPU, RAM, storage, or special accelerators such as GPUs. More information here (the description (documentation metadata) of the artifact(s)).
- Code Artifact Software Dependencies. Document the OS required, as well as any third-party libraries required to execute the experiments.
- Experimental Workflow. This is the *most important* step for the documentation of the artifact. In this section, authors include (i) a complete description of the experiment workflow that the code can execute, (ii) an estimation of the execution time to execute the experiment workflow, (iii) a complete description of the expected results and an evaluation of them, and most importantly (iv) how the expected results from the experiment workflow relate to the results found in the article. Best practices indicate that, to facilitate the understanding of the scope of the reproducibility, the expected results from the artifact should be in the same format as the ones in the article. For instance, when the results in the article are depicted in a graph figure, ideally, the execution of the code should provide a (similar) figure (there are open-source tools that can be used for that purpose such as gnuplot). It is critical that authors devote their efforts to these aspects of the reproducibility of experiments to minimize the time needed for their understanding and verification.
Steps for the AE phase – Full reproducibility evaluation
To minimize the effort required by reviewers and promote the future reuse of results, it is highly desirable to maximize automation throughout the entire process. The attainment of this goal can be facilitated by packaging the artifacts (such as with docker containers) and offering installation and deployment scripts.
- Data artifact availability. In case your computational artifacts include any kind of dataset, it must be made stored in an *open*, public, persistent repository, such as Zenodo. These open repositories assigned a Digital Object Identifier (What is a DOI? Check this out) that allow researchers to identify the dataset univocally.
- Data artifact documentation. Then, for the ICPP24, the dataset must be documented. It can include methodological aspects about how the dataset was obtained, if there was any preprocessing and transformation, etc. More information in the computational artifact(s).
- Code artifact Hardware dependencies. Specify and document hardware requirements (including type and quantity). It could be the case that the code artifact does not require any particular hardware requirements, then it should be stated. In some cases, the requirements can involve specific CPU, RAM, storage, or special accelerators such as GPUs. More information here (the description (documentation metadata) of the artifact(s)).
- Code Artifact Software Dependencies. Document the OS required, any third-party libraries required. If using container technologies, as recommended, the documentation of this step can be easily accomplished by looking at and the dockerfile.
- Experimental Workflow. This is the *most important* step for the documentation of the artifact. In this section, authors include (i) a complete description of the experiment workflow that the code can execute, (ii) an estimation of the execution time to execute the experiment workflow, (iii) a complete description of the expected results and an evaluation of them, and most importantly (iv) how the expected results from the experiment workflow relate to the results found in the article. Best practices indicate that, to facilitate the understanding of the scope of the reproducibility, the expected results from the artifact should be in the same format as the ones in the article. For instance, when the results in the article are depicted in a graph figure, ideally, the execution of the code should provide a (similar) figure (there are open-source tools that can be used for that purpose such as gnuplot). It is critical that authors devote their efforts to these aspects of the reproducibility of experiments to minimize the time needed for their understanding and verification, for instance, by stating how the execution outcome matches the experiments in the paper exactly and how long it takes to execute the experiments. Authors need to consider how long it is reasonable for reviewers and third-party researchers to wait for the experiments. For the cases of long run executions, it is advisable to provide an additional reduced-scale version of the experiment that can lead to analogous conclusions.
- Code Artifact availability. In case your computational artifacts include any type of code, the code must be stored in an *open*, public, persistent repository. We recommend using github that can be subsequently paired with Zenodo. Zenodo provides a DOI to the code, so that it guarantees that it cannot be modified while the reviewers are examining it, and in the future, when third-party researchers and practitioners are reusing it.
- Packaging Method of the Code Artifact. We strongly recommend container technologies to provide the code, which simplifies the deployment and installation processes and helps document the dependencies. More information about packaging methods here (The computational artifact(s))
- Installation & Deployment. Prepare installation and deployment scripts and document the installation and deployment process, including an estimation of the installation and deployment times. The documentation should also include (i) the process description to install and compile the libraries and the code, and (ii) the process description to deploy the code in the resources. The description of these processes should include an estimation of the installation, compilation, and deployment times. When any of these times exceed what is reasonable, authors should provide some way to alleviate the effort required by the potential recipients of the artifacts. For instance, capsules with the compiled code can be provided, or a simplified input dataset that reduces the overall experimental execution time. On the other hand, best practices indicate that, whenever it is possible, the actual code of software dependencies (libraries) should not be included in the artifact, but scripts should be provided to download them from a repository and perform the installation.
- Configuration of the installation / deployment of the experiment in ChameleonCloud. To alleviate the burden of reviewers and third-party researchers, we highly recommend authors to prepare their scripts for the ChameleonCloud infrastructure. More information in the computational resources required to enable reproducibility.